Gene name: porA.
Designation within complete genome sequence of strain CJ11168: Cj1259.
Location within CJ11168: Start/End 1189121 / 1190395.
Length within CJ11168: 1275bp / 424 aa.
Molecular weight: 45688 Da.
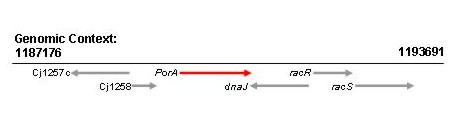
Location of the nucleotide sequence fragment used in the database (red) within the complete gene sequence
The PCR amplification primer binding sites are highlighted in blue. This nucleotide sequence (red) from strain 11168 has been assigned allele 34.
ATGAAACTAGTTAAACTTAGTTTAGTTGCAGCTCTTGCTGCAGGTGCTTTTTCAGCAGCT AACGCTACTCCACTTGAAGAAGCGATCAAAGATGTTGATGTATCAGGTGTATTAAGATAC AGATACGATACAGGTAATTTTGATAAAAATTTCGTTAACAACTCAAATTTAAACAACAGC AAACAAGATCACAAATATAGAGCACAAGTTAACTTCAGTGCTGCTATAGCTGATAACTTC AAAGCTTTTGTTCAATTTGACTATAATGCTGCTGATGGTGGTTATGGTGCTAATGGAATA AAAAATGATCAAAAAGGACTTTTTGTTCGTCAATTATACTTAACTTATACAAATGAAGAT GTTGCTACAAGTGTAATCGCTGGTAAACAACAATTAAACCTTATCTGGACGGATAACGCT ATTGATGGTTTAGTTGGCACAGGTGTTAAAGTAGTAAATAACAGCATCGATGGTTTAACT CTAGCTGCTTTTGCTGTAGATAGCTTCATGGCTGCAGAGCAAGGTGCAGATTTATTAGAA CATAGTAATATTTCAACAACATCAAATCAAGCTCCTTTTAAAGTAGATTCAGTAGGAAAT CTTTACGGTGCTGCTGCTGTAGGTTCTTATGATCTTGCTGGTGGACAATTCAACCCACAA TTATGGTTAGCTTATTGGGATCAAGTAGCATTCTTCTATGCTGTAGATGCAGCTTATAGT ACAACTATCTTTGATGGAATCAACTGGACACTTGAAGGTGCTTACTTAGGAAATAGCCTT GATAGCGAACTTGATGATAAAACACACGCTAATGGCAATTTATTTGCTTTAAAAGGTAGC ATTGAAGTAAATGGTTGGGATGCTAGCCTTGGTGGTTTATACTACGGTGATAAAGAAAAA GCTTCTACAGTTGTAATCGAAGATCAAGGTAATCTTGGTTCTTTACTTGCAGGTGAGGAA ATTTTCTATACTACTGGTTCAAGACTAAATGGTGATACTGGTAGAAATATCTTCGGTTAT GTAACTGGTGGATATACTTTCAACGAAACAGTTCGCGTTGGTGCTGACTTCGTATATGGT GGAACAAAAACAGAAGCTGCTAATCATTTAGGTGGTGGTAAAAAACTTGAAGCTGTTGCA AGAGTAGATTACAAATACTCTCCAAAACTTAACTTCTCAGCATTCTATTCTTATGTGAAC CTAGATCAAGGTGTAAACACTAATGAAAGTGCTGATCATAGCACTGTAAGACTTCAAGCT CTTTACAAATTCTAA
Location of the MOMP peptide fragment used in the database (red) within the intact protein
This peptide sequence from strain 11168 has been assigned allele 31
MKLVKLSLVAALAAGAFSAANATPLEEAIKDVDVSGVLRYRYDTGNFDKNFVNNSNLNNS
KQDHKYRAQVNFSAAIADNFKAFVQFDYNAADGGYGANGIKNDQKGLFVRQLYLTYTNED
VATSVIAGKQQLNLIWTDNAIDGLVGTGVKVVNNSIDGLTLAAFAVDSFMAAEQGADLLE
HSNISTTSNQAPFKVDSVGNLYGAAAVGSYDLAGGQFNPQLWLAYWDQVAFFYAVDAAYS
TTIFDGINWTLEGAYLGNSLDSELDDKTHANGNLFALKGSIEVNGWDASLGGLYYGDKEK
ASTVVIEDQGNLGSLLAGEEIFYTTGSRLNGDTGRNIFGYVTGGYTFNETVRVGADFVYG
GTKTEAANHLGGGKKLEAVARVDYKYSPKLNFSAFYSYVNLDQGVNTNESADHSTVRLQA
LYKF
Trimming sequences to the correct length for database queries
The porA alleles and MOMP peptides contained in this database are different lengths due to insertions and deletions within their sequences (see below). However, they have been trimmed to begin and end within highly conserved regions representing equivalent sequence positions.
Before querying the database, new sequences should be trimmed to the correct length. To identify the trim sites we recommend that the nucleotide sequence is translated and the amino acid sequence aligned as shown below. This assists the identification of the correct trim sites, which are residues DSF at the N-terminus and KKL at the C-terminus, within the yellow boxes.
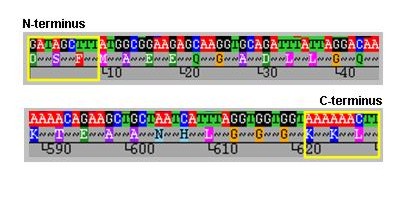
MOMP peptide sequences aligned from the N-terminal trimming site.
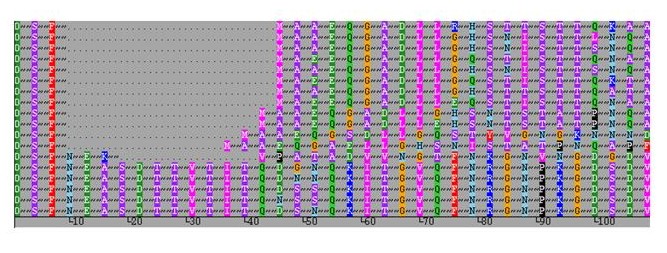
Occasionally a sequence / peptide motif may be repeated, as shown in the example below.
